Megamolecules
Our group pioneered a new platform for engineering protein-based molecules – called Megamolecules – rooted in meticulous synthetic biology and synthetic chemistry approaches. In short, we selectively react the enzyme component of recombinant fusion proteins with selective inhibitors at the end of synthetic chemical linkers to precisely assemble these large scaffolds. This approach has superior specificity and kinetics, often several orders of magnitude greater than traditional bioconjugation chemistries. Because of this distinct synthetic advantage, we can build molecules with much greater control and precision of their structure and complexity than with traditional protein engineering techniques, for example, structures composed of more than twenty protein domains, with branched, cyclic and dendritic architectures, and with molecular weights greater than one Megadalton. Currently, we are applying this platform for many applications, including dynamic protein structures, multifunctional molecules, and diagnostics and therapeutics for many health conditions.

Synthetic Chemistry
Linker Synthesis
Combining novel chemical synthesis methods with optimized enzymatic suicide inhibition, synthetic linker development allows us to expand the shape and size of our megamolecule toolbox.
Synthetic Biology
Building Block Assembly
Utilizing our published solid-phase synthesis platform to build our megamoleucle libraries, the inherently modular design allows us to probe any indication that sparks scientific interest and need.
Engineering
Application
Exploring large design spaces, we are identifying design rules for building protein-based molecules for a wide range of applications, including conformationally dynamic structures and diagnostic and therapeutic agents.
Linker Synthesis
Fundamentally, the reaction of recombinant proteins and synthetic linkers are based on the selective reaction of an electrophilic warhead with an active-site residue of an enzyme. This ensures that it is rapid and specific. Furthermore, many of these intermediates are stable in solution for periods of several days.
The two primary eletrophilic molecule-enzymes combinations that are used for long-term scaffold design are:
SnapTag, a mutant O6-alkylguanine DNA alkyltransferase that specifically reacts with chloro-pyrimidine groups.
Cutinase, a serine esterase that specifically reacts with ethyl p-nitrophenyl phosphonate ligands to give phosphonate esters.
Other enzmye-reactant pairs in lab have been investigated for short-term/programmed hydrolysis, including HaloTag (a-chloroalkanes) and Beta-lactamase.
Currently, we can covalently link these reacting groups at the end of a long hydrophilic poly(ethylene glycol) chains (linker synthesis), which allows us to then selectively construct megamolecule scaffolds that react with cutinase or SnapTag enzymes. Ideally, new, stable chemistries will develop, allowing for bioorthogonal reaction schemes that can expand the megamolecule toolbox into large, precise, and beautiful geometries. This will allow for even more expansive structure-function relationship investigation with therapeutics and diagnostics our group continues to develop.
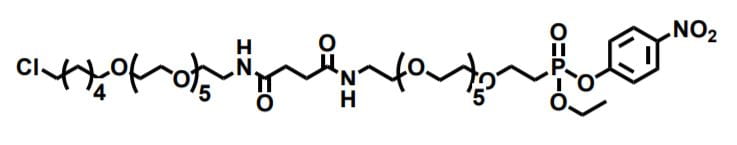
Figure 1. An example of a bispecific synthetic linker that reacts with HaloTag (left) and cutinase (right).
Building Block Assembly
As we have recently shown in our 2020 JACS communinication, Solid-Phase Synthesis of Megamolecules, we were able to draw inspiration from standard solid-phase peptide synthesis to precisely build megamoledule G2 dendrons around 200kDa. These molecules were so large that we are able to beautifully confirm their conformations and size (~25nm) by transmission electron microscopy (TEM).
From this success, we are able to delve into the construction of antibody-enzmye or antibody-drug conjugates for cancer therapy. Specific epitopes seen on the surface of cancer cells can be targeted by the antibody portion of these conjugates, while either the drug or enzyme + prodrug conversion can locally kill malignant cells. This method is truly powerful, as the homogeneous product allows for more meticulous structure-function relationships to be further investigated.
Application
In work supported by the Air Force Office of Scientific Research, we are synthesizing modular, printable megamolecule inks for incorporation in advanced nanoprinting technologies. In work supported by the U.S. Army Research Office, we are building protein-based molecular structures that are responsive to stimuli, providing dynamic control over their conformation. In work supported by many private and Northwestern Institutes, we are developing molecules that act as targeted diagnostics or therapeutics for cancer and other malignancies.
Megamolecules Publications:
Modica, J.A., Skarpathiotis, S., Mrksich, M. Modular Assembly of Protein Building Blocks to Create Precisely Defined Megamolecules. ChemBioChem. 13, 16, 2331–2334 (2012).
Modica, J.A., Lin, Y., Mrksich, M. Synthesis of Cyclic Megamolecules. J. Am. Chem. Soc. 140, 20, 6391–6399 (2018).
Taylor, E.L., Metcalf, K.J., Carlotti, B., Lai, C.-T., Modica, J.A., Schatz, G.C., Mrksich, M., & Goodson III, T. Long-Range Energy Transfer in Protein Megamolecules. J. Am. Chem. Soc. 140, 46, 15731–15743 (2018).
He, P., Zuchnairz, J., Zhou, S., Modica, J.A., Dhindwal, S., Li, Y., Voth, G.A., Mrksich, M., Dravid, V.P., Roux, B. Modeling Synthesized Protein Megamolecules: Sctructure, Dynamics, and Functions. Biophysical Journal. 118, 3, 517 (2020).
Kimmel, B.R., Modica, J.A., Parker, K., Dravid, V.P., Mrksich, M. Solid-Phase Synthesis of Megamolecules. J. Am. Chem. Soc. 142, 10, 4534-4538 (2020).
Modica, J.A., Iderzorig, T., Mrksich, M. Design and Synthesis of Megamolecule Mimics of a Therapeutic Antibody. J. Am. Chem. Soc. 142, 13657-13661 (2020).
Metcalf, K.J., Kimmel, B.R., Sykora D.J., Modica, J.A., Parker, K.A., Berens, E., Dai, R., Dravid, V.P., Werb, Z., and Mrksich, M. Synthetic Tuning of Domain Stoichiometry in Nanobody-Enzyme Mega-molecules. Bioconjugate Chem. 32, 143-152 (2021).
